News & Topics
ホーム > Research > News & Topics > Multiclass logistic regression was applied on dementia-miRNome data from NCGG Biobank
In a new paper published in Scientific Reports
published in Scientific Reports on 22nd Oct, researchers at the Medical Genome Center, Research Institute, NCGG have proposed a dementia subtype prediction model by multiclass classification using blood-based microRNA (miRNA) biomarkers. Dr. Yuya Asanomi, researcher and lead author on the paper, and his colleagues constructed the prediction model with 46 miRNA biomarkers by using miRNome (comprehensive miRNA expression levels) data stored in NCGG Biobank
on 22nd Oct, researchers at the Medical Genome Center, Research Institute, NCGG have proposed a dementia subtype prediction model by multiclass classification using blood-based microRNA (miRNA) biomarkers. Dr. Yuya Asanomi, researcher and lead author on the paper, and his colleagues constructed the prediction model with 46 miRNA biomarkers by using miRNome (comprehensive miRNA expression levels) data stored in NCGG Biobank .
.
Biomarkers discovery for dementia is extremely difficult. There are various types of dementia, and the differential diagnosis is important for appropriate care and treatment. The development of novel biomarkers for differentiating dementia subtypes is much needed. In this study, a multiclass logistic regression was applied to the serum miRNA expression data to construct a model for dementia subtype prediction. NCGG Biobank stores miRNome data for approximately 5,000 individuals including dementia patients. The miRNA expression profile from ~1,300 dementia patients (Alzheimer's disease, vascular dementia, dementia with Lewy bodies, and normal pressure hydrocephalus) and ~250 normal cognitive individuals were used in this study. The prediction model was constructed based on penalized regression methods for multiclass classification. The optimal number of miRNAs and the penalty parameter were determined through cross-validation in a training group, and the final prediction model was constructed using the entire training group data. Finally, a prediction model using 46 miRNA biomarkers was obtained, and the model classified the validation group into four subtypes of dementia.
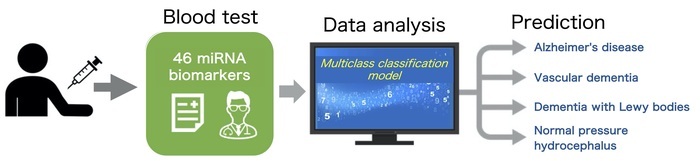
Differential diagnosis by multiclass classification using blood-miRNA biomarkers
The prediction accuracy of the model constructed in this study was not sufficient yet. However, the potential of multiclass classification using blood-based miRNA biomarkers was demonstrated, and further investigation using larger sample sizes will improve the prediction accuracy. Additionally, this method might be applicable to the stratification of other complex diseases. Furthermore, the network analysis of the genes targeted by the 46 identified miRNAs was performed. Two genes associated with Alzheimer's disease pathogenesis, SRC and CHD3, were detected as hub genes in the target gene network of the model for Alzheimer's disease. This suggests that the miRNA markers used in this model are suitable for the prediction of the dementia subtypes.
Title: Dementia subtype prediction models constructed by penalized regression methods for multiclass classification using serum microRNA expression data
Authors: Yuya Asanomi, Daichi Shigemizu, Shintaro Akiyama, Takashi Sakurai, Kouichi Ozaki, Takahiro Ochiya, Shumpei Niida
Journal: Scientific Reports